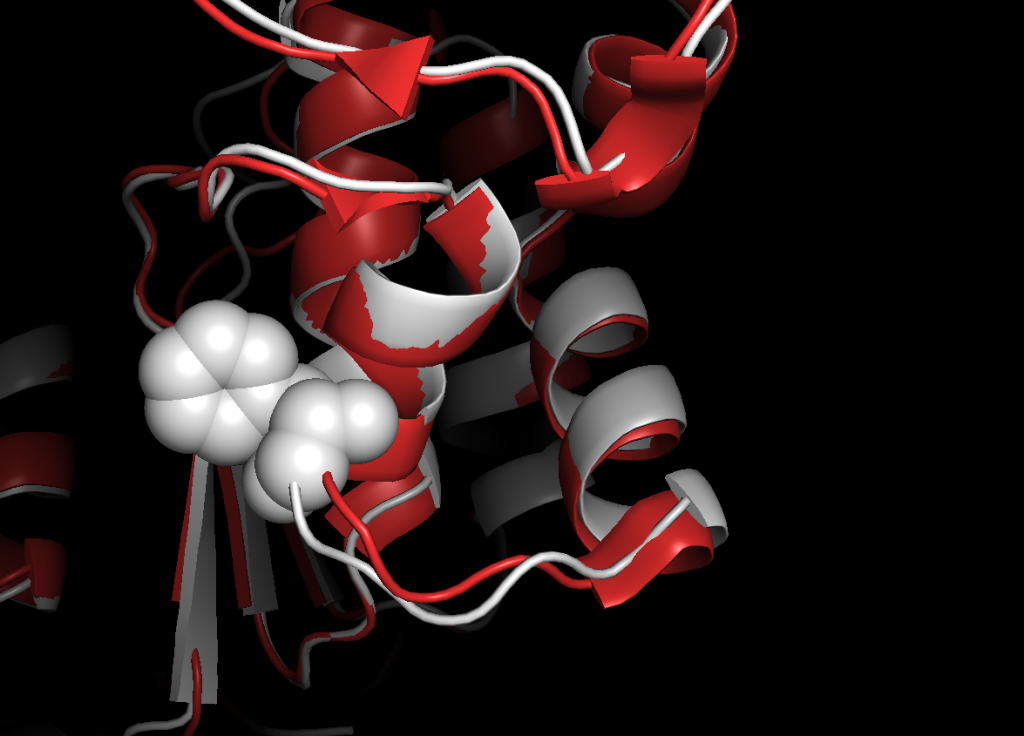
This weekend I implemented a protein aligner which can overlay proteins such that they can be compared (nothing new, standard technique). I overlayed a normal with a mutated CFTR-NBD1-region (PDB 1XMJ with F508del mutation, PDB 2BBO the wildtype sequence). See the result below. The white protein is the wildtype (normal) sequence and the red one being the one with the mutation. The white spheres are the Phenylalanine which is missing in the mutated version.
I was surprised by the similarity. The variation between the two looks to be non-significant for the function of the protein and I recall to have read that in fact the F508del-mutatant protein does it’s job almost as well as the wildtype (but is usually faster recycled if it gets to the cell surface). The problem with the F508del mutation is the breakdown of the protein before reaching the membrane, because the endoplasmatic reticulum discards it due to failing quality checks (details are currently fuzzy, sorry, citations needed). Several approaches to rescue the mutant protein are currently being investigated, most prominently the Orkambi® medicine, which already is available but is combined in new ways to improve function (https://www.biorxiv.org/content/early/2018/05/11/320630), and microRNA-techniques (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5488273/).
But when there is no misfolding of the protein, it must be discarded on it’s sequence, which makes me (without having reviewed the literature on that topic) hypothesize that there are mechanisms in the cell (somewhere in the folding pathway) that detect peptide sequences as foreign and mark proteins for lysis, similar to the way antibodies outside the cell detect foreign substances. My next focus will hence become to see what peptide sequences are naturally occurring in humans and which not and might hence be detected as foreign. As a kick-off I searched the peptide sequences of the GRCh38-DNA reference sequence-project for sequences similar to the wildtype and the mutated sequences. I found none other in the forward direction, but found the 5-3-terminal-inverted sequence VGIINE (the sequence around Phe508 being ENII(F)GV) in the TRPV1-protein. What the significance of this is and how similar the structures are is my next task.
Comments are closed.